Introduction
Fermentation is a phenomenon in which nutrients such as carbohydrates are decomposed anaerobically by microorganisms, and in a broad sense, the overall production of useful substances by culturing microorganisms is also called fermentation. It is known through many studies that active ingredients that exhibit superior functions than natural extracts or raw materials are produced through a microbial fermentation process that decomposes them using enzymes produced by microorganisms compared to the extracted natural ingredients or raw materials themselves (Bae et al., 2019).
Fermented seafood, such as salted seafood, is aged by various decomposing enzyme reactions secreted by microorganisms with the addition of salt, and it has been reported that a large number of bacteria exhibiting salt resistance grow (Park et al., 2017), and the isolated halotolerant microorganisms are derived from various natural environments with high salinity content and are being applied to various industrial fields such as phylogenetic diversity, as well as the development of industrially applicable halotolerant enzymes, and treatment or recycling of aquatic by-products (Singh et al., 2016).
Proteolytic enzymes, which account for the largest proportion in the industrial enzyme market, are used in food, detergents, cosmetics, and pharmaceuticals (Singh et al., 2016), and proteolytic enzymes as a starter to shorten the aging period of fermented seafood. Active research is being conducted, such as analysis of the community of halotolerant microorganisms that produce (Giyatmi and Irianto, 2017;Zang et al., 2020). In addition, proteolytic enzymes produced by halotolerant microorganisms can be used to decompose the shells of aquatic products generated in the aquaculture industry to produce animal collagen peptides as hydrolysates, so proteolytic enzymes can achieve a high added value of waste resources. Alternatively, methods using microorganisms that produce them are being studied (Wu et al., 2008;Mayuri et al., 2019). In addition, microorganisms for salted foods can grow in acidic or alkalic conditions as well as high salt concentrations, and those with strong proteolytic activity are more likely to be used.
However, hydrolytic enzymes produced by general microorganisms are biocatalysts and have the advantage of being more eco-friendly than chemical reactions catalyzed by chemical substances. In order to solve this problem, it is thought that it will be possible to solve the problem by using a hydrolytic enzyme derived from a microorganism that can grow even under high salt concentrations, acidic or alkalic conditions.
Therefore, in this study, halotolerant microorganisms were isolated from freshwater samples from two Nakdong River estuary wetland (Samrak Wetland Ecological Garden and Daejeo Ecological Park), and three representative hydrolytic enzymes (amylase, lipase, protease) activity, and various hydrolase production characteristics were investigated along with the search for the cultural characteristics of the isolated microorganisms. Analysis of auxin production capacity to confirm the possibility of use as a microbial fertilizer in the eco-friendly agriculture field by having the function of promoting the growth of crops from what was reported (Jung et al., 2006;Jung et al., 2007) was also carried out. This is a study on the search for halotolerant microbial resources derived from domestic wetland freshwater, and it is expected to enhance the value of the isolated strains and to be used as a basic biological material for bioengineering research related to hydrolytic enzymes for the food and cosmetic industries.
Materials and Methods
Screening of the halotolerant bacteria was done from the water samples of freshwater wetlands from Samrak Wetland Ecological Garden (35°10'25.8"N 128°58'21.9"E) and Daejeo Ecological Park (35°11'57.7"N 128°58'35.9"E), two representative wetlands of the Nakdong River Estuary in Korea. Isolation of halotolerant bacteria was done by serial diluting each sample in 0.85% saline buffer 10-1 to 10-4 and aerobically cultivating the plates at 37 °C using Marine agar medium (BD, New Jersey, USA). After cultivation, the same solid medium was used for further single colony isolation of halotolerant bacteria based on the morphological properties of the colonies. Pure colonies were obtained after several transfers, and the growth of the isolated bacteria at 37 °C on complex media were tested using Nutrient agar (BD, New Jersey, USA), R2A agar (BD, New Jersey, USA), and tryptic soy agar (BD, New Jersey, USA). As a halotolerant bacteria, the growth in the presence of 5-10% sodium chloride concentration was tested using marine agar medium, and optimal condition was confirmed. Also, optimum pH condition was confirmed using the same medium prepared with different pH at 5, 7, 9.
For bacterial identification, strains isolated from domestic wetland freshwater samples under aerobic conditions were cultivated on marine agar medium, and plates were sent to BIOFACT Co., Ltd. for 16S rDNA sequencing. Taxonomic identification based on the 16S rDNA sequence of the strains was analyzed using 16S-based ID service in EzBioCloud Database (https://www.ezbiocloud.net/) developed by Chun- Lab Inc. Phylogenetic analysis was done using BioEdit and MEGA X program.
Hydrolytic enzyme (amylase, lipase, protease) activities of the isolated halotolerant bacteria were tested on a selective agar medium supplemented with specific substrates to react accordingly. Hydrolytic enzyme activity was identified by cultivating the spot inoculated plates with halotolerant isolates using marine agar medium supplemented with 0.2% soluble starch (BD, New Jersey, USA), 1% Tween 80 (Sigma-Aldrich, Massachusetts, USA), and 2% skim milk (BD, New Jersey, USA) for amylase, lipase, protease activity, respectively. After 7 days of incubation at 37 °C, the plates were examined for enzyme activities by size of the clear zone around colonies. The area of the clear zone was determined as total area minus colony area (+++: > 7 mm, ++: 4-6 mm, +: 1-3 mm).
The auxin activity assay was performed using Salkowski reagent after incubating the isolates at 37 °C for 5 days in marine broth (BD, New Jersey, USA) supplemented with 0.1% L-tryptophan (Sigma-Aldrich, Massachusetts, USA). 800 μl of Salkowski reagent (35% HClO4, 2% FeCl3) was added to 400 μl culture supernatant and incubated in the dark for 30 mins. Auxin activity was measured by the change of the color (Red: ++; Pale red: +; Orange: w; Colorless: -).
Results and Discussion
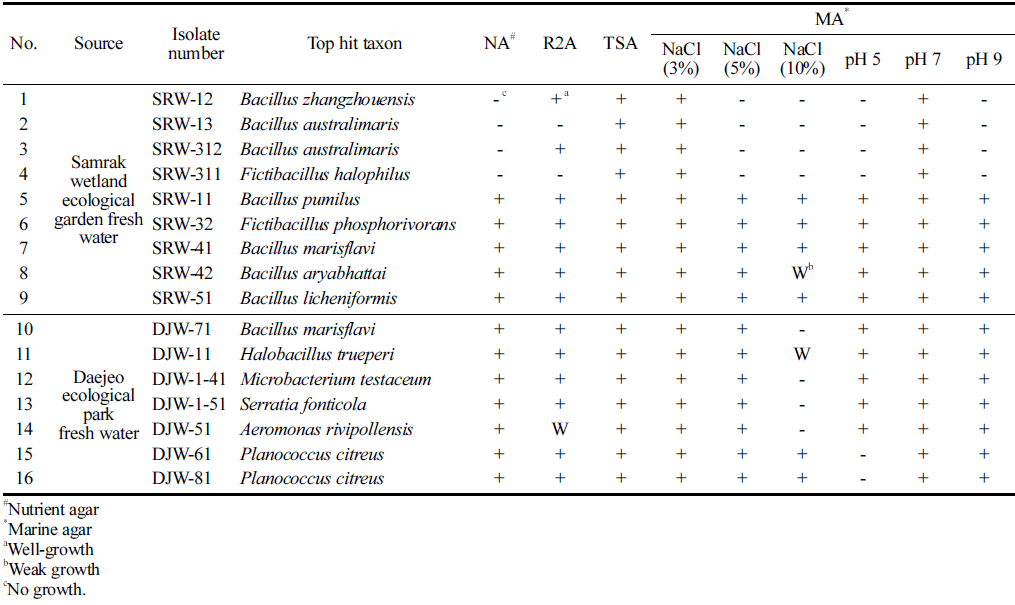
Halotolerant bacteria with possible industrial capability for food and cosmeceutical market was isolated from domestic Nakdong River estuary wetland freshwater samples (Samrak Wetland Ecological Garden and Daejeo Ecological Park wetland freshwater) aerobically culturing the samples using marine agar medium. Pure single colonies were obtained by streak plate method based on their morphological chracteristics. Screening result, as shown in Table 1, total of 16 strains of halotolerant microorganisms were isolated, 9 strains from the freshwater sample of Samrak Wetland Ecology Park, and 7 strains were isolated from Daejeo Ecological Park through this work. In addition, the marine agar medium, mainly composed of mineral salts, is a favorable medium for the cultivation of seawater-derived microorganisms. Confirmation of the potential industrial application of the strains was tested using complex media (nutrient agar, R2A agar, tryptic soy agar) that are being used for mass cultivation of the microorganisms. As a result, all 16 strains were able to grow (including weak growth) in at least one type of complex medium. Salt tolerance of the halotolerant strains was tested at 5% and 10% salt concentrations using marine agar medium (MA, 3% NaCl). As a result, it was confirmed that 8 strains (50%) were able to grow (including weak growth) under both 5% and 10% NaCl conditions, and 4 strains (25%) grew only under 3% NaCl conditions. Also, in order to confirm optimal pH conditions for growth, a marine agar medium adjusted at pH 5, 7, 9 was used. All isolated strains were able to grow at pH 7, and 10 strains (No. 5, 6, 7, 8, 9, 10, 11, 12, 13, 14) showed growth at pH 5 and pH 9 (Table 1).
|
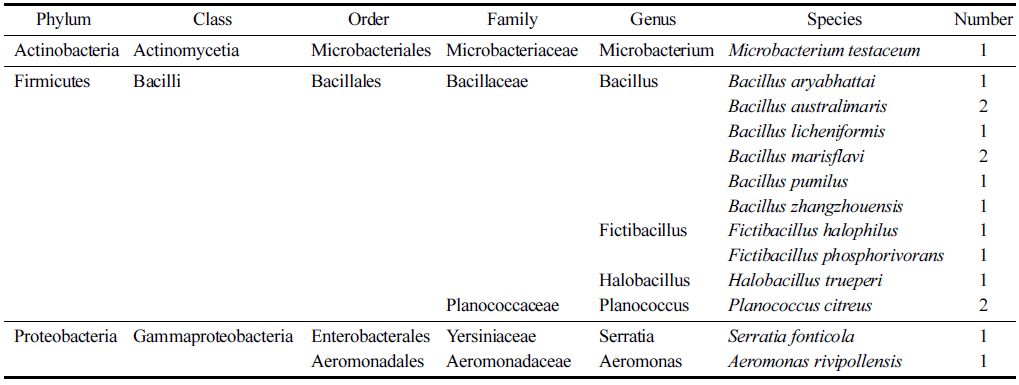
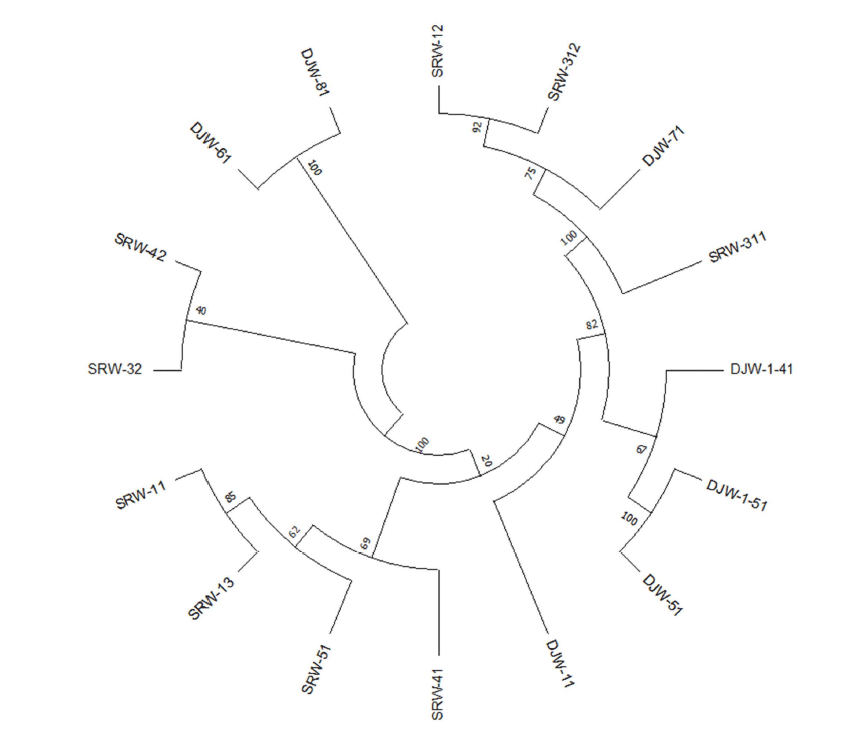
The results of microbial identification using EzBioCloud's 16S-based ID analysis service on the basis of 16S rDNA nucleotide sequence of 16 aerobically isolated strains from domestic wetland freshwater samples were largely divided into 3 phyla, 3 classes, 4 orders, 5 families, 7 genera, and 13 species (Table 2), and the identification result of the isolated strains and their closest strains definition is shown in Table 3. In addition, a phylogenetic tree was constructed to confirm the relationship between the isolated and identified strains (Fig. 1). As shown in Table 2, Firmicutes (Bacilli) was dominant with 81.3%, Proteobacteria (Gammaproteobacteria) was next with 12.5%, and Actinobacteria (Actinomycetia) was the least with 6.2%. The 13 strains from phylum Firmicutes that dominated were distributed in two families Bacillaceae and Planoco- ccaceae, with 84.6% and 15.4%, respectively. From phylum Proteobacteria, only Gammaproteobacteria class species were isolated, and isolated strains belonged to the family Yersiniaceae and Aeromonaceae, each accounting 50%. Lastly, 1 strain belonged to the family Microbacteriaceae in the phylum Actinobacteria.
|
|
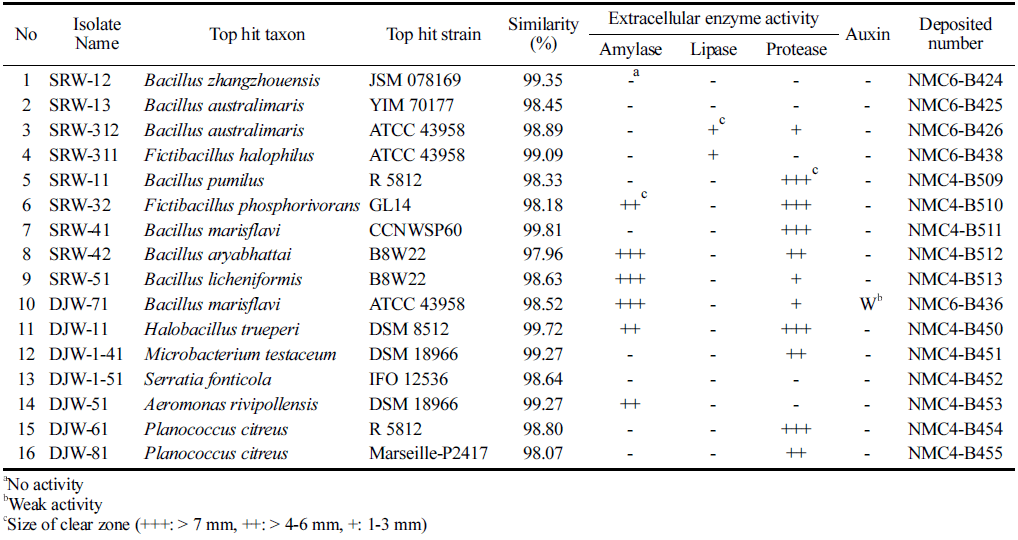
Analysis of hydrolytic enzymes and auxin activity was performed for possible application of isolated halotolerant strains in the food and cosmeceutical industries and potential use as microbial fertilizer in the eco-friendly agricultural industry. As shown in Table 3, it was confirmed that all strains, except 3 strains, had at least one hydrolytic enzyme activity and 5 strains (Deposit number: NMC4-B510, NMC4-B512, NMC4-B513, NMC6-B436 & NMC4-B450) showed more than 2 enzyme activities. Weak auxin production was confirmed in 1 strain (Deposit number: NMC6-B436) isolated from the freshwater sample of Daejeo Ecological Park.
Duan and their colleagues conducted a characteristic study of β-amylases produced by the Bacillus aryabhattai GEL-09 strain and reported that they could be applied for starch hydrolysis (Duan et al., 2021). Therefore, it is expected to be applied to the food industry of Bacillus aryabhattai (Deposited number: NMC4-B512), which is one of the isolated strains with high amylase and protease activity in this study. Moreover, the antibacterial ability of Bacillus licheniformis SCK B11 against harmful bacteria and the characteristics of biogenic amine decomposition were reported (Kim et al., 2012), indicating that Bacillus licheniformis (Deposited number: NMC4-B513) also can be used as a candidate for manufacturing of traditionally fermented soybean products that are difficult to control pathogens and biogenic amine levels.
In addition, the halotolerant microorganisms isolated through this study will be of great significance in terms of securing the diversity of domestic microbial biological resources. The eight isolates from the wetland showed growth at 10% NaCl concentration and these isolates showed growth at pH 9 unlike other isolates from the same wetland sample. These 4 isolates (NMC4-B509, NMC4-B510, NMC4-B511 & NMC4-B454) showed highest protease activity corresponding to +++ (Table 1 & 3). Alkaline protease from these halotolerant strains that grow under high salt concentration conditions can be used not only for detergents but in food-grade products broadening its application range (Mayuri et al., 2019). Therefore, in this study, auxin production capacity analysis was carried out for use as a microbial fertilizer that replaces chemical fertilizers by promoting the growth of crops along with the activity of three enzymes (amylase, lipase, protease) applicable to starch processing, food industry, and industrial enzyme market for detergent using halophilic microorganisms isolated from domestic wetlands located close to the sea. It is expected that this will enhance the value of halophilic microbial resources derived from wetlands and be used as basic biological materials for industrial enzyme-related biotechnology and microbial fertilizer research (Jeong et al., 2021).
It is expected that it can be used as a basic biological material for enzyme-related biotechnology for application to the food and cosmetic industries as well as the possibility of discovering new strains. Also, all strains isolated through this study were deposited in the Microbial Value Enhancement Project, Korea Research Institute of Bioscience and Biotechnology.
Summary
This study investigated the cultural characteristics and enzyme productivity of halotolerant microorganisms isolated from the wetland samples of the Nakdong River Estuary in Korea. The isolation of halotolerant bacteria was carried out by cultivating aerobically at 37 °C using a marine agar medium. After incubation, 16 pure single colonies were obtained, and phylogenetic analysis was done based on the 16S rRNA sequence using the 16S-based ID analysis program in the EzBioCloud database showing the strains were divided into 3 phyla, 5 families, 7 genera, and 13 species. Also, in order to confirm the industrial applicability of the isolated strains, the enzyme activity was evaluated to confirm the culture conditions, and the production of industrially valuable enzymes such as amylase, lipase, and protease were performed. It was confirmed that 10 strains were able to grow in the pH range between pH 5 and pH 9, and 8 strains were able to grow under 10% NaCl conditions. 13 strains had at least one enzyme activity, and 5 strains showed activity of two enzymes. This indicates the possibility of using the microorganisms isolated through this study in the food and cosmetic industries. Therefore, in this study, it is thought that the use of purely isolated strains through this study will be helpful in securing domestic genetic resources and improving strains to improve hydrolytic enzyme production capacity.







