Introduction
Currently, approximately 400 species of lactic acid bacteria (LAB) have been documented, and are typically classified into seven genera including Leuconostoc, Oenococcus, Pediococcus, Lactobacillus, Enterococcus, Streptococcus, and Lactococcus (Carr et al., 2002; Salminen et al., 2004; Zhang et al., 2011). Among these genera, Leuconostoc comprises 24 different species (Kot et al., 2014). This genus contains non-motile, gram-positive, and catalase-negative facultative anaerobes, with DNA exhibiting a relatively low GC content (37-45 mol%). Leuconostoc is associated with meat, poultry, fish, raw milk, dairy products, and various materials of plant-origin (Kim & Chun, 2005; Björkroth & Holzapfel, 2006). It has been determined that different species of Leuconostoc play an important role in fermenting foods such as cheese, kimchi, and sauerkraut. Among LAB, Leuconostoc citreum is one of the most established bacterial strains used for manufacturing kimchi (a Korean traditional dish). It is listed in the generally recognized as safe (GRAS) category; therefore, it plays an important role in various applications in pharmaceutical and biotechnology industries. One important and promising application is the use of these bacteria as a host for the delivery of therapeutic proteins into humans or animals (Son et al., 2016). The accurate identification of LAB is required for both basic research and food-based industrial applications (Tanigawa & Watanabe, 2011). For the rapid identification and analysis of microbial communities, classical biochemical methods can be used, which involve assessing morphological, physiological, and biochemical properties for preliminary identification of bacteria from different sources. However, these techniques are not adequately efficient and sometimes, results can be inconclusive (Tafvizi & Ebrahimi, 2015). Therefore, correct identification and classification of LAB is difficult without the support of genotypic techniques (Kandler & Weiss, 1986; Gevers et al., 2001). Currently, various molecular typing methods such as random amplified polymorphic DNA (RAPD), restriction fragment length polymorphisms (RFLP), pulse-field gel electrophoresis (PFGE), protein fingerprinting, and repetitive element palindromic PCR (rep-PCR) are available for the identification and characterization of LAB species (Villani et al., 1997; Cibik et al., 2000; Pérez et al., 2002; Sánchez et al., 2005; Björkroth & Holzapfel, 2006; Vihavainen & Björkroth, 2009; Nieto-Arribas et al., 2010; Alegría et al., 2013). Genetic fingerprinting techniques can be used to characterize single bacterial isolates or bacterial communities. It was previously published that LAB genotypes can be identified by partial 16S rRNA gene sequencing followed by characterization using species specific PCR (Holzapfel et al., 2001). Among PCR-based methods, RAPD has been used for the differentiation and classification of LAB (Drake et al., 1996; Cocconcelli et al., 1997; Quiberoni et al., 1998; Tailliez et al., 1998; Cibik et al., 2000; Cardamone et al., 2011). The objective of this study was to examine natural LAB populations associated with different food sources in Korea and to report the molecular identification and characterization of 14 bacterial strains.
Materials and Methods
Fourteen bacterial strains were isolated from seven different food sources, specifically, kimchi (6), salted small octopus (3), pear (1), young radish kimchi (1), salted oyster (1), salted fish (1), and sweet pumpkin (1) from Korea. Strains were grown overnight on MRS (Difco Laboratories, Detroit, MI, USA) plates at 30°C. For long-term preservation, isolated cell suspensions were stored in broth cultures supplemented with 1.5% (w/v) glycerol at -80°C.
For DNA isolation, bacterial cells from 3 mL overnight cultures were pelleted by centrifugation at 16,000×g for 1 min and the pellet was washed with 1 mL of TE buffer (0.1 M Tris- HCl, 0.01 M EDTA, 1 M NaCl). Subsequently, genomic DNA was isolated using an AccuPrep Genomic DNA Extraction kit (Bioneer, Daejeon, Korea) following the manufacturer’s procedure. The quality of DNA was validated using a 0.8% (w/v) agarose gel and 1× TAE buffer.
All isolated strains were identified using a set of two universal primers, specifically, 27F (5'-AGA GTT TGA TCM TGG CTC AG-3') and 1492R (5'-TAC GGY TAC CTT GTT ACG ACT T-3') for amplifying the 16S rRNA gene. The PCR mixtures were subjected to the following thermal cycling conditions: 94°C for 30 s, annealing at 50°C for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 7 min, in a 20 μL reaction volume, using the Bioneer Premix kit. Amplified products were analyzed using 0.8% agarose gel electrophoresis and purified using a Promega Kit (Gel and PCR cleanup system, Promega, WI, USA) following the manufacturer’s instructions. The purified DNA was sequenced using an ABI 3730 xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA) and service from Macrogen sequencing (Macrogen Inc., Seoul, Korea). The 16S rRNA gene sequence homology was compared using the NCBI database with BLASTN.
RAPD primers for this study were commercially synthesized by Macrogen (Macrogen Inc.). Two universal primers, 239 (5'- CTG AAG CGG A-3') and KAY3 (5'-CTG GCG ACT G-3'), which are usually used for the taxonomic discrimination of lactic acid bacteria, were used at a concentration of 10 pmol/ μL for RAPD-PCR typing of the 14 isolated bacterial strains. For PCR amplification, 1 μL of purified genomic DNA, 2 μL of each primer (for two separate reactions), and 17 μL of distilled water were added to the premix kit (Bioneer - 10 mM Tris-HCl (pH 9.0), 40 mM KCl, 1.5 mM MgCl2, 250 μM dNTP mix, and 1 U of Taq polymerase) to a final volume of 20 μL. In an automatic thermocycler (My Cycler, Bio-Rad, CA, USA), the reaction mixtures were subjected to the following temperature cycles: 94°C for 4 min, 2 cycles of denaturation at 94°C for 2 min each, annealing at 39°C for 2 min, extension at 72°C for 2 min, 35 cycles of denaturation at 95°C for 15 s each, annealing at 39°C for 15 s, and extension at 72°C for 20 s, followed by a final elongation at 72°C for 7 min. For electrophoresis, 5 μL of each amplified PCR product was loaded onto a 1.2% (w/v) agarose gel (Lonza, Rockland, NY, USA) in 1× TAE buffer. For DNA staining, 0.5 μL of the ethidium bromide solution (10 mg/mL) was directly added to each 10 mL of agarose solution. Two molecular ladders of 100 bp (Bioneer) and 1 kb (Takara Bio Inc., Shiga, Japan) were used as size markers. After electrophoresis, the gel was photographed using a gel documentation system (BioRad Gel Doc XR+) to verify the PCR results. Finally, the band pattern was analyzed using the Quantity One software and phylogenic trees were constructed by the unweighted pair group method with arithmetic mean (UPGMA) method.
Results and Discussion
The objective of this study was to develop a simple, costeffective, easy to understand, RAPD-based molecular tool for typing bacterial strains isolated from different food sources from Korea. For the identification of isolated bacteria, 16S rRNA gene sequencing was performed. Currently, 16S rRNA gene sequencing is the most commonly used method to study bacterial phylogeny and taxonomy. This is possibly because the 16S rRNA gene is present in all bacteria (Woese, 1987). BLAST analysis confirmed that all 14 isolated strains were Leuconostoc citreum. The gene sequences have been submitted to GenBank with the accession numbers (KX286339- KX286352). It has been demonstrated that the use of traditional diagnostic testing (i.e. sugar fermentation patterns) alone, commonly leads to misidentifications among Leuconostoc and other closely related genera such as Lactobacillus, Weissella, and Pediococcus (Kulwichit et al., 2007).
DNA-based molecular techniques have been confirmed as valuable tools to assess the identity and diversity of different strains (Aznar & Alarcon, 2002). The PCR-based RAPD typing technique uses 10-15 bp primers that bind to multiple genomic sites to detect sequence diversity. The main advantage of this method is that it is fast, easy to perform, and inexpensive, and it has high discriminatory power (Pozo- Bayon et al., 2009). In a previous study (Cibik et al., 2000), a combination of RAPD-PCR and 16S rDNA sequencing with specific primers was applied to analyze the molecular diversity of a large collection of Leuconostoc strains mainly isolated from traditional French cheeses.
In the present work, a preliminary taxonomic study was performed using 14 isolates from different food sources from Korea. Genomic DNA of 14 L. citreum strains was successfully amplified, and the results obtained using two oligonucleotide primers (239 and KAY3) and RAPD analysis are illustrated in Fig. 1. In total, 130 bands were obtained and seven different RAPD profiles were produced using both primers. The RAPD profile with primer 239 consisted of eight major bands of 2000, 1600, 1200, 1000, 600, 480, 350, and 200 bp (Fig. 1(A)), whereas with primer KAY3, seven major bands of 4000, 2500, 2000, 1600, 1000, 600, and 500 bp were reported (Fig. 1(B)). Many of the strains shared common DNA band sizes with both primers (for example, strains 11037, 11082, 11382, 11391, 11413, 11414, and 11422 with primer 239 and strains 11037, 11076, 11082, 11093, 11382, 11414, and 11447 with primer KAY3) as expected, because all strains were initially identified as L. citreum. Furthermore, with primer 239, the 2000 bp band was only present in strain 11001, and the 500 bp and 600 bp bands were unique to strains 11093 and 11388, respectively. The 350 bp band was prominent in only one sample (11432); similarly, the 200 bp band was identified in two strains (11076 and 11447) of the 14 selected for this study. In contrast to primer 239, a unique band of 500 bp was observed with strain 11388 using primer KAY3 (Fig. 1(B)). The maximum band size observed here was 4000 bp in sample 11432, but this band was very faint. The RAPD typing method has been used for the identification and discrimination of several LAB strains of Pediococcus, Lactobacillus, Enterococcus, and Oenococcus among others (Corroler et al., 1998; Nigatu et al., 1998; Morea et al., 1999; Mora et al., 2000; Suzzi et al., 2000; De Angelis et al., 2001; Moschetti et al., 2001). There has been an increasing need to select microbial strains with commercial applications and to improve quality- and safetyrelated aspects of traditional fermented foods (Fortina et al., 2003).
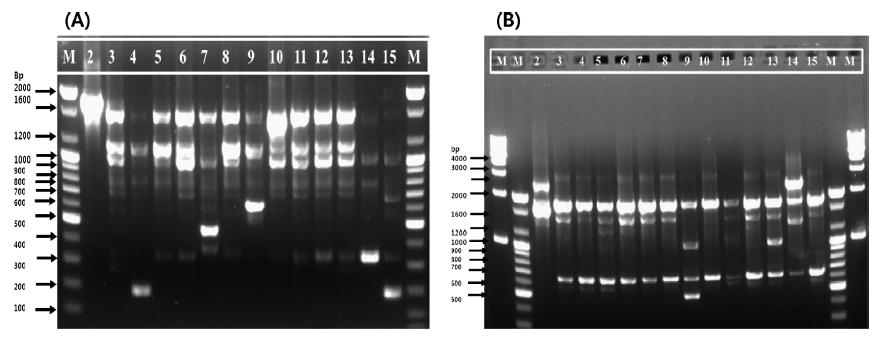
Strains with similar RAPD-PCR patterns should not be regarded as identical, even when they come from the same sources, because they might possess dissimilar industrial and/ or biological attributes (Cardamone et al., 2011). To understand the phylogenetic relatedness between isolates, a dendrogram was constructed using the Quantity One software. Dendrogrambased analysis with primer 239 showed that all 14 strains could be divided into seven clades out of which clade VII had the maximum of seven (11382, 11077, 11037, 11082, 11414, 11422, and 11413) strains of 14. Clade V had only two strains (11388 and 11076). Clades I, II, III, IV, and VI were composed of one strain each (11001, 11447, 11391, 11432, and 11093, respectively), as shown in Fig. 2(A). In addition, a single representative sample from each clade revealed a unique banding pattern (Fig. 1(A)). In contrast, dendrogram analysis with the primer KAY3 divided the 14 L. citreum strains into four clades out of which clade IV consisted of a maximum of 10 (11447, 11382, 11413, 11037, 11093, 11082, 11391, 11077, 11414, and 11076) strains of 14. Clade I was represented by two strains (11432 and 11001) (Fig. 2(B)). Clades II and III were composed of strains 11388 and 11422, respectively. Samples in lanes 2, 14, 9, and 13 were representative of clades I, II, and III, which were separated from the major group and showed different RAPD gel band patterns in comparison to the other samples, using primer KAY3 (Fig. 1(B)). It was observed that some isolates grouped together with one primer, but changed groups with another primer. It has been suggested that the problem of RAPD-PCR reproducibility can be overcome by using rigid laboratory protocols. Ramos et al. (2008) developed a procedure to optimize repeatability and avoid bias in sampling loci for genetic analyses based on RAPD data.
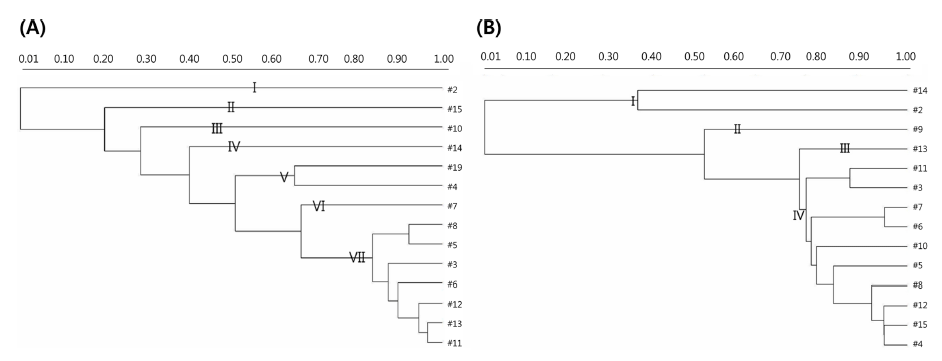
In addition, a single typing method is not sufficient for the identification of bacteria from different food sources. It can be concluded that a multidimensional approach using biochemical characterization, 16S rRNA gene sequencing, and different molecular typing methods could lead to accurate species identification.
The current study presents the phylogenetic relationships and genetic variations among L. citreum strains isolated from different food products in Korea. RAPD profiles are useful for preliminary screening and typing of bacterial isolates at a lower cost and higher speed. Isolation of novel strains for industrial applications is of utmost importance. For identification of such novel strains, other molecular typing methods are also required. The use of such novel strains in the food industry could offer great potential for augmenting existing processes or for developing new applications.







