Introduction
Proteus mirabilis, often isolated from the gastrointestinal tract, is a member of Enterobacteriaceae (Daly et al., 2016) and a pathogenic species related to human diseases that can cause nosocomial infections. P. mirabilis can cause various infections in humans, such as in the skin, wounds, eyes, ears, nose, respiratory tract, and gastrointestinal tract and, in particular, has been reported as a urinary tract infectioncausing bacterium (Jacobsen & Shirtliff, 2011).
Recently, interest in the relationship between intestinal imbalance and Parkinson’s disease (PD) has been increasing. PD is a progressive neurological disorder that exhibits symptoms such as rigidity, resting tremors, slowness of movements, and postural instability. As a neuropathological characteristic of PD, dopaminergic neurons can be lost in the substantia nigra pars compacta (Hirsch et al., 2005). Reactive nitrogen species play a pathogenic role in the brains of patients with PD by modifying α-synuclein and parkin. Protein tyrosine residues or free tyrosine react with nitrating oxidants and produce 3-nitrotyrosine, a marker for reactive nitrogen species. Reactive nitrogen species induce dopaminergic neuron death (Blanchard-Fillion et al., 2006). Thus, the accumulation of α-synuclein and 3-nitrotyrosine may induce apoptotic cell death. Furthermore, increasing intestinal penetrability, lipopolysaccharide-binding protein in plasma, colonic inflammation, and pro-inflammatory cytokines, such as tumor necrosis factor-α and interleukin-1β, were reported as causes of PD (Devos et al., 2013;Kelly et al., 2014;Pellegrini et al., 2016). P. mirabilis is also related to Crohn’s disease (CD) and can cause inflammation in cells and animal models of colitis in the gut (Manichanh et al., 2006). Commensal flora has various abilities to induce inflammation and protective immune responses (Sartor, 2001). Changes in the composition of intestinal bacteria and the abundance and diversity of reduced microorganisms were observed in patients with CD (Manichanh et al., 2006).
Lactic acid bacteria (LAB) are classified as Gram-positive, spore non-forming, anaerobic, non-motile, and rod- or coccusshaped bacteria that can be found in various sources. LAB are important in food fermentation as they can ferment carbohydrates and produce lactic acid if supplied into the medium. Enterococcus, Lactobacillus, Lactococcus, Leuconostoc, Pediococcus, and Streptococcus are representative genera found in a variety of fermented foods (Nettles & Barefoot, 1993). Probiotic LAB have interesting therapeutic properties, such as improving the gut immune system and biological availability of nutrients and reducing lactose intolerance and formation of pathogenic biofilms from pathogenic bacteria (Gómez et al., 2016). LAB produce various metabolites from glucose fermentation. Some LAB produce antimicrobial compounds as metabolites, including low-molecular-mass compounds such as organic acids (lactic, acetic, formic, propionic, and butyric acids), ethanol, fatty acids, acetoin, hydrogen peroxide, carbon dioxide, diacetyl, antifungal compound, bacteriocins, and bacteriocin-like high-molecularmass compounds (Reis et al., 2012;Yang, 2000).
Antibiotics are chemicals that prevent bacterial growth by stopping the cell from dividing or killing bacterial cells. Antibiotic-resistant bacterial infections, such as respiratory infections and skin and tissue infections, are some of the most serious challenges and are now widespread in both healthcare settings and the wider community (Lushniak, 2014). Antibioticresistant infections increase patient morbidity and mortality as antibiotic treatments are not effective against them (Weiner et al., 2016). As the incidence of drug-resistant pathogens increases, natural antibacterial agents are attracting attention as an alternative strategy to prevent infectious diseases (Savoia, 2012).
In this study, a Weissella cibaria strain with high antimicrobial and antibiofilm activities against P. mirabilis was isolated from kimchi. The antimicrobial compounds were partially purified from the culture supernatant of the selected strain, and their antimicrobial activity was evaluated.
Materials and Methods
A total of 987 LAB strains from our laboratory were used. Proteus mirabilis KCCM 11381P provided by the Korean Culture Center of Microorganisms (KCCM; Seoul, South Korea) was also used. The LAB strains were cultured in lactobacilli de Man, Rogosa, and Sharpe broth (MRS; BD, Franklin Lakes, NJ, USA), while P. mirabilis was cultured in nutrient broth (BD) and brain heart infusion broth (BHI; BD).
Pathogenic strains and LAB strains obtained from our laboratory stock (Korean Culture Collection of Probiotics, KCCP), KCCM, Korean Collection for Type Cultures (KCTC; Jeongup, South Korea), and the American Type Culture Collection (ATCC; Manassas, VA, USA) were also used. Escherichia coli ATCC 10536 was cultured in Luria-Bertani broth (BD), Bacillus cereus KCCM 11204, Staphylococcus aureus KCCP 60043, Salmonella Enteritidis ATCC 13076, Pseudomonas aeruginosa ATCC 10145, Salmonella Typhimurium ATCC 14028, and Listeria monocytogenes KCCM 40307 were cultured in nutrient broth, Weissella paramesenteroides KCCP 11717 and Leuconostoc lactis KCCP 11247 were cultured in MRS broth, and Candida albicans KCTC 7270 was cultured in yeast malt broth (BD). All strains were incubated at 37 °C.
The LAB strains were cultured on MRS agar plates and used to prepare the culture supernatant. After incubating a single colony in 1 mL of MRS at 37 °C for 20 h, the culture supernatant was collected by centrifugation at 14,000×g for 5 min to obtain the 1× supernatant. The prepared culture supernatant was used to examine the growth inhibition activity against P. mirabilis.
Antimicrobial activity was measured using a modified version of the paper disc test, also known as the disc diffusion method (Pulusani et al., 1979). A paper disc (diameter 6 mm; ADVANTEC CO., LTD., Tokyo, Japan) loaded with 15 μL of the culture supernatant of LAB was placed onto an agar plate overlaid with P. mirabilis (1.5 × 108 CFU/mL). After incubation at 37 °C for 24 h, the diameter of the inhibition zone surrounding the paper disc was measured to evaluate the antibacterial activity of the supernatant of the LAB strain. Any strain that exhibited a large inhibition zone was considered as having high antibacterial activity.
As described by Cho et al. (2023), 16S rRNA sequencing and phylogenetic analyses were carried out. Genomic DNA was extracted using the AccuPrep ® Genomic DNA Extraction Kit (Bioneer, Daejeon, South Korea), and the 16S rRNA gene was amplified through polymerase chain reaction (PCR) using AccuPower ® PCR PreMix (Bioneer) and primers. The nucleotide sequences of the 16S rRNA gene were determined by Bionics Co., Ltd. (Seoul, South Korea) and analyzed using BLAST from NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic trees were constructed using ClustalX (Larkin et al., 2007) and MEGA (Saitou & Nei, 1987) software.
Gram staining and catalase test were carried out as described by Cappuccino & Sherman (1983). The selected LAB strains were observed under a scanning electron microscope (SEM; H-7600, Hitachi, Tokyo, Japan) installed at Eulji University (Seongnam, South Korea). The selected sample was prepared by fixing it with glutaraldehyde and osmium tetroxide, dehydrating it with ethanol, and coating it with platinum, following the methods of Cho et al. (2023) and Kang et al. (2023). ImageJ software (Schneider et al., 2012) was used for the size analysis of scanning electron micrographs. The utilization pattern of carbohydrates was determined using the API 50 CHL kit (API System, bioMérieux, Montalieu Vercie, France) according to the manufacturer’s instructions.
To concentrate the culture supernatant, 1 mL of 1× supernatant was processed using a speed vac (VC2127, LaboGene A/S, Lynge, Denmark) to obtain 100 μL of 10× supernatant. Freeze-dried supernatant was also concentrated using a freeze dryer (FDU-1110, EYELA, Tokyo, Japan). The freeze-dried supernatant was dissolved in 20 mM sodium phosphate buffer (pH 7.0) to a final concentration of 1 g/mL.
To determine the optimal incubation time for the production of antimicrobial compounds, 1× supernatant was collected at specific time points (0, 3, 6, 9, 12, and 24 h) during incubation. The antimicrobial activity of each sample was measured using the disc diffusion method.
To determine the stability of the antimicrobial compound at different pH levels, the pH of the 1× supernatant was adjusted to 2, 3, 4, 5, 6, 7, 8, 9, and 10 using 1 N NaOH (Samchun Chemical Co., Ltd., Seoul, South Korea) and 1 N HCl (Samchun Chemical Co., Ltd.) Each supernatant was incubated at 30 °C for 3 h, and then the pH was adjusted to 6.5 with 1 N NaOH or 1 N HCl. The antimicrobial activity of each supernatant was evaluated using the disc diffusion method.
A 1× supernatant of the strains was exposed to a range of temperatures: 40, 50, 60, 70, 80, 90, and 100 °C for 10 min. The antimicrobial activity of the supernatant after exposure to different temperatures was evaluated using the disc diffusion method.
Proteinase K (Sigma-Aldrich, Burlington, MA, USA), trypsin (Roche, Basel, Switzerland), and α-chymotrypsin (Sigma- Aldrich) were used. Each enzyme was added to 1× supernatant at a final concentration of 1 mg/mL and incubated at 37 °C for 3 h. The effect of the enzymes on the antimicrobial activity of the supernatant was measured using the disc diffusion method.
The antimicrobial compounds were partially purified using gel permeation column chromatography as described by Huttunen et al. (1995) with slight modification. Glass Econo- Column (1.5 × 120 cm) (Bio-Rad, Hercules, CA, USA) was packed with Bio-Gel P-2 polyacrylamide gel (Bio-Rad) and pre-equilibrated with distilled water. Freeze-dried supernatant was loaded onto the column and eluted at a rate of 0.3 mL/min. Eluents were fractionated with a volume of 3 mL using a fraction collector (Bio-Rad) and 25-fold concentrated using a speed vac. The antimicrobial activity of each fraction was evaluated using the disc diffusion method, and the active fractions, which showed antimicrobial activity, were pooled.
The antimicrobial activity of the selected fractions was confirmed through thin-layer chromatography (TLC). Each fraction was spotted onto TLC plates (silica gel 60F254, Merck, Darmstadt, Germany). The plates were then developed twice with a mobile phase consisting of nitromethane, 1- propanol, and water in a ratio of 2:5:1.5 (v/v/v) (Seo et al., 2004). The developed plate was dried, and spots of antimicrobial compounds were visualized by spraying with a solution of 0.3% (w/v) N-(1-naphthyl) ethylenediamine dihydrochloride (Sigma-Aldrich) in methanol and 5% (v/v) H2SO4 (Duksan, Seoul, South Korea), followed by heating at 120 °C for 3 min. The glucose polymers (G1-G8, Carbosynth Co., Compton, UK) were used as standards.
The concentration of reducing sugar and protein in a sample was determined using the Somogyi-Nelson method (Nelson, 1944;Somogyi, 1952) and Bradford protein assay (Bradford, 1976), respectively.
The molecular size of antimicrobial compounds was determined using size exclusion high-performance liquid chromatography (HPLC, UltiMateTM 3000 RSSC nanosystem, Thermo Fisher Scientific Inc., Waltham, MA, USA). A Shodex OHpak SB-802.5 column (8.0 × 300 mm, Resonac Co., Tokyo, Japan) was used. HPLC-grade distilled water was used as the mobile phase, and the flow rate was 0.4 mL/min. The column oven temperature was set at 35 °C, and a refractive index detector (Shodex RI-101, Resonac Co.) was used. Glucose polymers (G1, G2, G3, G5, and G7) (Carbosynth Co.) were used as standards.
The antibiofilm activity of a set of antimicrobial compounds was evaluated using a modified crystal violet assay (described by Lee et al., 2021). Briefly, W. cibaria strains were cultured in MRS at 37 °C for 20 h, and their culture supernatant was diluted 2-fold (1/2, 1/4, and 1/8) with phosphate-buffered saline.
To assess the effect of the culture supernatant of W. cibaria strains on the biofilm formation, P. mirabilis was cultured in BHI broth at 37 °C for 20 h. The BHI broth, which was inoculated with P. mirabilis culture at a concentration of 1% (v/v), was then mixed with the diluted culture supernatant of W. cibaria strains at a 1:1 ratio (v/v) and incubated in a 96-well plate at 37 °C for 48 h to allow for biofilm formation. Fresh MRS broth was used as a control instead of the diluted culture supernatant of W. cibaria strains.
To assess the effect of the culture supernatant of W. cibaria strains on the removal of preformed biofilm, P. mirabilis was cultured in BHI broth in a 96-well plate at 37 °C for 48 h to allow for biofilm formation. The 96-well plate was carefully washed twice with phosphate-buffered saline to remove any planktonic bacteria. The preformed biofilm was then incubated with the culture supernatant of the W. cibaria strain at 37 °C for 24 h. The pre-formed biofilm incubated with fresh MRS broth was used as a control instead of being incubated with the culture supernatant of the W. cibaria strain.
After the biofilm was stained with crystal violet, as described by Yoo (2022), the absorbance of the sample was measured at a wavelength of 595 nm using a microplate reader (Epoch microplate reader, BioTek Instruments Inc., Winooski, VT, USA). The inhibition rate of biofilm formation and removal rate of preformed biofilm of the sample were calculated using the following formula:
where Ac represents the absorbance of the well treated with the culture supernatant of W. cibaria strain, and A0 represents the absorbance of the control.
The experiments were conducted in triplicate, and the data are presented as mean±standard deviation. Statistical analysis was performed using SPSS 23 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA). One-way analysis of variance (ANOVA) followed by Duncan’s multiple range test (p<0.05) was used to determine the significance between groups.
Results and Discussion
A total of 987 LAB strains from our laboratory were used to screen for strains with high growth inhibitory activity against Proteus mirabilis. Among 987 LAB examined, 32 strains had growth inhibition zone of 10 mm or more in diameter, and only one strain, CHK 903, had a growth inhibition zone of over 15 mm in diameter. Strain no. CHK903, which was isolated from kimchi, showed an inhibition zone of 16 mm and, therefore, was selected as the strain with the highest antimicrobial activity against P. mirabilis for this study.
Bartkiene et al. (2019) reported that Lactobacillus brevis strain no. 173 exhibited an inhibition zone of 15.3 mm against P. mirabilis. Additionally, LAB (Leuconostoc, Lactobacillus, Enterocococus, and Pediococcus species) isolated from sourdough have also been found to possess antibacterial activity against P. mirabilis. In another study, W. confusa isolated from a vaginal swab from Indian women showed antimicrobial activity against P. mirabilis (15 mm) (Purkayastha et al., 2017). These reports and our results indicate that various types of LAB have high antimicrobial activity against P. mirabilis and may serve as potential antibacterial agents against P. mirabilis.
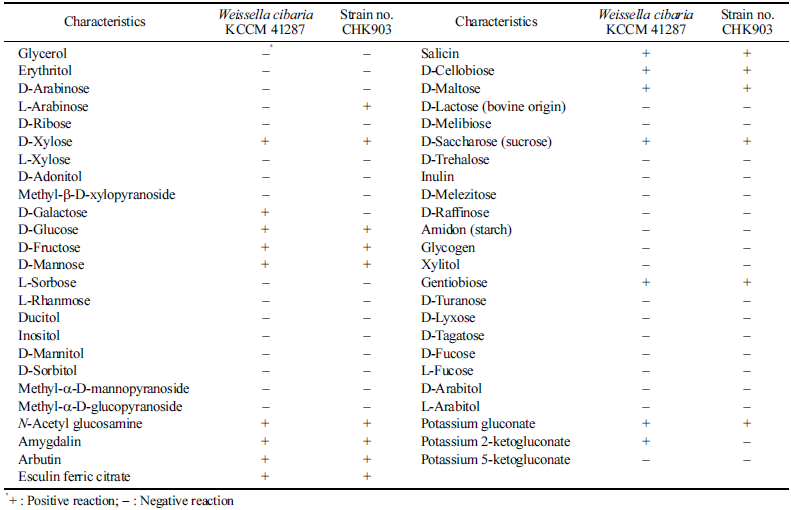
When the nucleotide sequence of the 16S rRNA gene of stain no. CHK903 was analyzed by BLAST, it was 100% identical to that of 87 strains of W. cibaria and to that of four strains of W. confusa registered in the NCBI database, which confirms the genus Weissella (data not shown). The selected strain was identified as W. cibaria through phylogenetic tree analysis (Fig. 1a), and SEM analysis showed that the length of the cells ranged from 1.59 to 1.93 μm, while the width ranged from 0.68 to 0.80 μm (Fig. 1b). The strain was rod-shaped with rounded ends and was Gram-positive and catalasenegative. These characteristics are consistent with the profile of LAB, which are typically cocci- or bacilli-shaped, Grampositive, and catalase-negative. The carbohydrate utilization pattern of W. cibaria CHK903 was determined and compared with that of the type strain W. cibaria KCCM 41287. W. cibaria CHK903 was confirmed to be able to ferment various carbohydrates (Table 1), such as L-arabinose, D-xylose, Dglucose, D-fructose, D-mannose, N-acetyl glucosamine, amygdalin, arbutin, esculin ferric citrate, salicin, D-cellobiose, D-maltose, D-saccharose, gentiobiose, and potassium gluconate. However, this strain could not utilize D-galactose, unlike the type strain. The results of the phenotypic and biochemical tests were consistent with the identification of the selected strain as W. cibaria, which was previously reported by Rao & Goyal (2013).

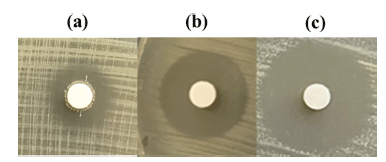
The inhibition zone diameter for W. cibaria CHK903 at 1× supernatant concentration was 15.3 mm (Fig. 2). When the supernatant was concentrated to 10×, the diameter of the inhibition zone increased by 1.5-fold, and when freeze-dried, it increased by 1.7-fold. This result indicates that the concentrated and freeze-dried supernatants had higher antimicrobial activity than the 1× supernatant. The pH of the supernatant from the selected strain was 4.6. At this pH, the diameter of the inhibition zone was 14.7 mm. After adjusting the pH to 6.5, the diameter of the inhibition zone slightly decreased (data not shown).
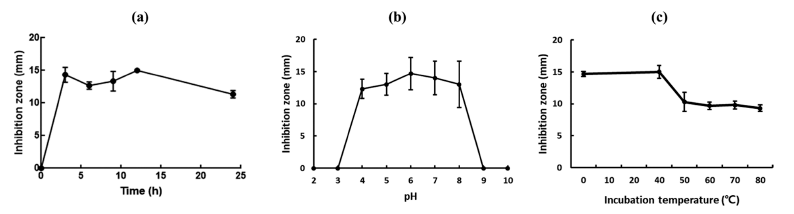
When the antimicrobial activity of the culture supernatant of W. cibaria CHK903 was determined at different time points (3, 6, 9, 12, and 24 h), the maximum antimicrobial activity of the strain was observed at 12 h after incubation (Fig. 3a). The antimicrobial activity of the supernatant between 3 and 12 h was similar but slightly decreased after 12 h of incubation.
The antimicrobial activity of the bacteriocin produced by B. cereus GN105 was found to be dependent on the growth phase. The antimicrobial activity was not observed during the exponential phase and decreased after 3 h during the stationary phase. Specifically, the antimicrobial activity was observed after 1.5 h and decreased after 7 h of incubation (Naclerio et al., 1993).
The stability of the antimicrobial activity of the culture supernatant of W. cibaria CHK903 at different pH levels is presented in Fig. 3b. The culture supernatant maintained its activity at pH 4 to 8 but completely lost its antimicrobial activity below pH 3 and above pH 9. Previous studies have shown that bacteriocins produced by L. brevis OG1 and Lactiplantibacillus plantarum F1 were stable at pH 2 to 8 and pH 2 to 6, respectively (Ogunbanwo et al., 2003). Compared to the stability of bacteriocins and antimicrobial compounds from other sources, the antimicrobial activity of W. cibaria CHK903 was found to be stable within a range of pH values.
Regarding temperature, the antimicrobial activity of W. cibaria CHK903 was maintained at 100% until 40 °C but decreased to 70% at 50 °C (Fig. 3c). Bacteriocin produced by B. cereus GN105 maintained its activity at temperatures up to 75 °C but lost activity after 15 min of exposure to 90 °C (Naclerio et al., 1993).
The culture supernatant of W. cibaria CHK903 was treated with proteolytic enzymes, such as proteinase K, trypsin, and α-chymotrypsin, and its antimicrobial activity was checked to determine whether the antimicrobial compounds are proteinaceous substances. As shown in Table 2, the antimicrobial activity of this strain was not affected by treatment with the proteolytic enzymes, which indicated that the antimicrobial compounds are not proteinaceous. LAB produce various proteins that have antimicrobial activity against pathogenic bacteria, such as bacteriocins. Bacteriocins are antimicrobial peptides or proteins produced by bacteria. They are typically small in size and have specific antimicrobial activity against related or unrelated bacteria (Ahmad et al., 2017). The results of the present study imply that the antimicrobial compounds produced by W. cibaria CHK903 are unlikely to be bacteriocin. Besides bacteriocins, the antimicrobial components of LAB are diverse and include organic acids, hydrogen peroxide, diacetyl, and other volatile organic compounds (Yang, 2000). Organic acids such as lactic, acetic, propionic, and formic acids are commonly produced by LAB during fermentation. These organic acids lower the pH of the environment and disrupt the proton motive force of bacteria, leading to cell death. Hydrogen peroxide (H2O2) is produced by some strains of LAB and generates reactive oxygen species that can damage bacterial DNA and other cellular components, leading to cell death (Reis et al., 2012). While there are some reports that lactic acid bacteria inhibit P. mirabilis growth, there is no report identifying the novel substances produced by lactic acid bacteria that inhibit P. mirabilis growth.

|
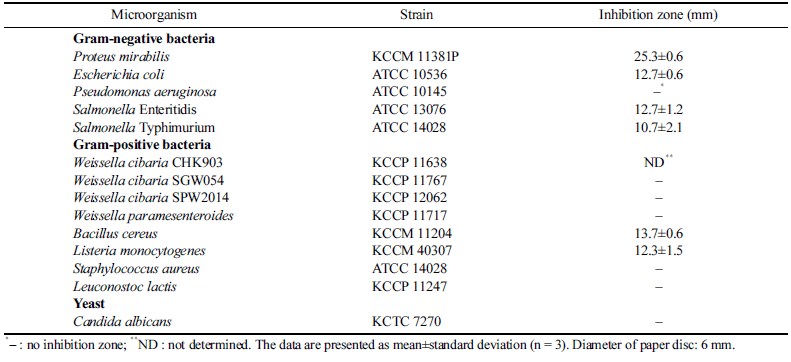
The antimicrobial activity of W. cibaria CHK903 against some pathogenic microorganisms and LAB was checked to evaluate the antimicrobial spectrum. As shown in Table 3, the strain inhibited the growth of E. coli, Salmonella Enteritidis, Salmonella Typhimurium, B. cereus, and L. monocytogenes. However, the strain could not inhibit the growth of W. cibaria, W. paramesenteroides, and Leuconostoc lactis. This result indicates that W. cibaria CHK903 selectively inhibits the growth of pathogenic bacteria and does not show antimicrobial activity against LAB. Previous studies have also reported significant inhibitory effects of W. cibaria against various bacterial strains, such as Acinetobacter baumannii, E. coli, L. monocytogenes, P. mirabilis, Salmonella Enteritidis, Salmonella Typhimurium, Staphylococcus epidermidis, and Streptococcus pyogenes (Yu et al., 2018;Jha et al., 2022). These findings suggest that W. cibaria CHK903 has the potential to serve as a natural food preservative in the food industry.
|
In this study, we revealed that the culture supernatant of W. cibaria CHK903 has high antimicrobial activity against P. mirabilis and some other pathogenic bacteria, including foodborne pathogens. Thus, the antimicrobial compounds from W. cibaria CHK903 were partially purified using gel permeation column chromatography. Among the fractions obtained from the eluent, samples with fraction numbers 37 to 38 and 33 to 36 showed antimicrobial activity with the average diameter of the inhibition zone being 14.5 and 12.5 mm, respectively. These results revealed that there were two antimicrobial compounds in the partially purified supernatant, which was also confirmed by TLC (Fig. 4a, b).
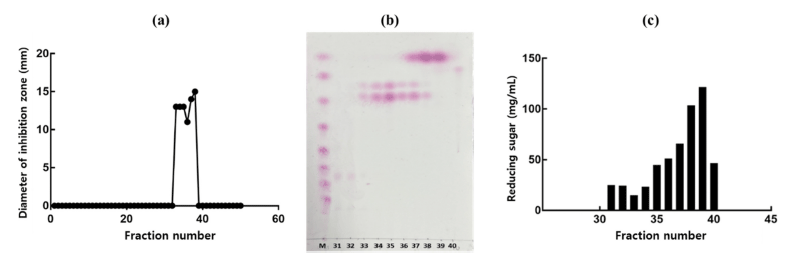
When the content of protein and carbohydrate of the partially purified antimicrobial compounds was determined to identify the antimicrobial compounds, it was confirmed that there was no protein in the sample, whereas reducing sugar was present, ranging from 14.2 to 24.3 mg/mL for fractions 31 to 34, 44.1 to 64.9 mg/mL for fractions 35 to 37, 102.9 and 120.9 mg/mL for fractions 38 and 39, and 46.1 mg/mL for fraction 40 (Fig. 4c). Based on these results, the active fractions were considered to contain carbohydrates and not protein.
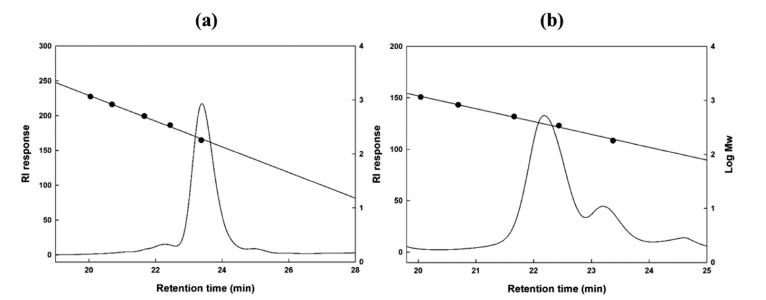
TLC analysis revealed two separate spots related to the antimicrobial compounds between glucose polymer G2 and G3 (Fig. 4b). The molecular weight of the two antimicrobial compounds was determined to be 189 and 365 Da (Fig. 5).
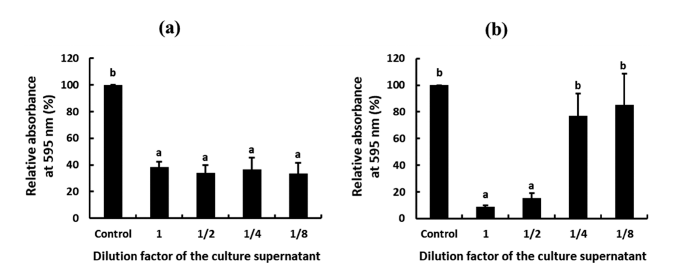
Besides the antimicrobial activity of W. cibaria CHK903 against P. mirabilis, the antibiofilm activity of the strain against P. mirabilis was evaluated. The antibiofilm activity can be divided into the inhibitory activity of biofilm formation and the removal activity of pre-formed biofilm. The supernatant of W. cibaria CHK903 showed dose-dependent inhibitory activity against the biofilm formation of P. mirabilis with an inhibition rate of 90% (Fig. 6a). The strain also exhibited removal activity against the pre-formed biofilm of P. mirabilis with a removal rate of over 60% (Fig. 6b). Biofilms are communities of microorganisms that adhere to biopolymers on surfaces, such as medical devices, and are composed of a selfproduced extracellular polymeric matrix. The matrix can reduce sensitivity to host defense systems, antibiotics, and other drugs, making biofilms a major contributor to chronic infections (Yan et al., 2019).
The results of our study suggest that the supernatant of W. cibaria CHK903 has significant potential and can be used as a natural antimicrobial and anti-biofilm agent against P. mirabilis. These properties make this strain valuable to various industries, including the food and medical fields, as its application could potentially prevent chronic infections and antibiotic resistance.
In conclusion, in this study, we evaluated the antimicrobial and antibiofilm activities of W. cibaria CHK903 against P. mirabilis. The selected strain exhibited potent antibacterial activity against P. mirabilis. The strains also showed broadspectrum antibacterial activity against Gram-positive and Gram-negative bacteria, including major foodborne pathogens. The partially purified antimicrobial compounds from W. cibaria CHK903 were identified as two compounds containing carbohydrates with molecular weights of 189 and 365 Da. However, the antimicrobial compounds partially purified in this study are heat-labile unlike carbohydrates, and it can be inferred that these compounds have additional residues bound to carbohydrates. This study also demonstrated the antibiofilm properties of W. cibaria CHK903, that is, its ability to inhibit biofilm formation and remove pre-formed biofilms of P. mirabilis. Given that P. mirabilis is associated with diseases such as urinary tract infections, pneumonia, otitis media, and PD, the findings in this study suggest that W. cibaria CHK903 has significant potential as a natural antimicrobial and antibiofilm agent for use in the prevention and treatment of diseases related to P. mirabilis.
Overall, this study provides valuable insights into the potential applications of W. cibaria CHK903 for developing natural antimicrobial agents and functional foods. Further research is needed to identify the antimicrobial compounds, elucidate their definite structure, explore the mechanism of antimicrobial activity, and develop effective formulations of these antimicrobial compounds for practical applications.