Introduction
Various environmental ecosystems are an interesting source of novel and promising microorganisms responsible for producing biocatalysts with great relevance in industrial areas. Soil, one of the understudied environments, is a fascinating source of microbial community (Sousa et al., 2022). Furthermore, soil microorganisms are known to produce diverse extracellular enzymes that degrade the complex of organic matter and are considered as the decomposers responsible for carbon recycling (Gomez et al., 2020).
Peat is the surface layer of soil that consists of organic matter and mineral water. The organic components of peat consist of water-soluble matter, organic solvent-soluble compounds, humus, lignin, and cutin (Jinming and Xuehui, 2011). Peat moss is the fibrous product of sphagnum moss and other organic materials that decompose in peat bogs over thousands of years. Due to the degradation of biomass, manure, peat, as well as compost, introduced into the fields as organic fertilizers, are considered as promising sources of mesophilic and thermophilic bacteria (Biyada et al., 2021).
Microorganisms based on their temperature range, in general, are classified as psychrophilic (0-20 ° C), mesophilic (20-45 ° C), and thermophilic (45-80 ° C), and the organisms that could grow above (+80 ° C) are referred as hyperthermophilic (Madigan et al., 2006). Although thermophiles are mainly found in hot springs and other geothermally heated systems, studies of a wide range of mesothermal environments have revealed that thermophiles are not confined to sites with high temperatures. The distribution of thermophiles in a mesothermal environment has been unintelligible (Wilson et al., 2022). The thermostable microorganisms were found to be at lower temperatures, with broader ranges, indicating the adaptation of these microorganisms to the wide temperature in various environments (Pandey et al., 2015).
In recent years, researchers have increasingly focused on the rich microorganism-based diversity of various environmental habitats to explore novel sources of enzymes that can perform under harsh industrial conditions. Hydrolases account for over 75% of all enzymes produced on a commercial scale. Among them amylases, lipases, and proteases are predominantly used in industrial bioprocesses. Approximately 24% of industrial enzymes are produced from bacteria (Fasim et al., 2021). For example, thermostable enzymes from bacteria are used in the production process of sweeteners, tagatose, and allulose, of CJ CheilJedang and Samyang Genex Corporation (Choi et al., 2016).
Thermal stability is a key characteristic of enzymes in numerous industrial biotechnological processes (Zheng et al., 2019). Enzymes are frequently exposed to an environment that differs from their natural environment during industrial processes (Littlechild et al., 2015). Thermophilic enzymes are thermally stable and have great catalytic activity at high temperatures, but their activity at moderate temperatures is lower than that of mesophilic enzymes. Thermophilic enzymes with improved low-temperature activity while maintaining high stability could be valuable in industrial processes. Mesophilic enzymes, on the other hand, have more catalytic activity than their thermophilic counterparts but are less thermally stable (Akanuma et al., 2019). Many protein engineering techniques have been developed to improve the thermal properties of mesophilic enzymes (Zheng et al., 2019). Apart from this, investigating the diversity of microorganisms, especially focusing on screening the mesophiles and thermophiles with a wide temperature range, could be an important consideration in ecological as well as biotechnological applications. In this study, the diversity and biotechnological potential of thermophilic and mesophilic microorganisms isolated from arable soil fertilized with peat moss were investigated.
Materials and Methods
The sampling site is located in Silla Garden in Busan, South Korea (35 ° 09'52.2"N 129 ° 00'02.2"E). Arable soil samples fertilized with peat moss from the West Siberian were collected from the surface layer (0-10 cm depth) in June 2017. Screening of bacteria was done using Marine broth (BD, Sparks, MD, USA) supplemented with 1.5% agar following the serial dilution technique. 1 g of soil sample was diluted in a 9 mL saline buffer and soil suspension was homogenized by vortexing. Diluted samples were inoculated into Marine agar (MA) plates and incubated at 37 ° C, 45 ° C, 50 ° C for several days. Subsequently, after incubation bacterial colonies growing on each diluted plate were transferred into the freshly prepared same solid medium. Pure colonies obtained after several transfers were determined for their physiological characteristics. The cultivation dependence of isolated bacteria on complex media was tested using Nutrient agar (BD, Sparks, MD, USA), R2A agar (BD, Sparks, MD, USA), and Tryptic soy agar (BD, Sparks, MD, USA). Growth temperature ranges of the isolates were determined by cultivating the isolates at 20 ° C, 25 ° C, 30 ° C, 35 ° C, 40 ° C, 45 ° C, 50 ° C, 55 ° C, 60 ° C, and 65 ° C. Growth pH range of the isolates were examined on MA medium adjusted at different pH of 4.0, 7.0, and 9.0. Salinity tolerance was determined using MA supplemented with NaCl (w/v) concentrations at 5% and 10%.
The identification was performed at the BIOFACT Co., Ltd using their standard pipeline. The 16S small subunit ribosomal gene was amplified with primers 27F, AGAGTTTGATCC TGGCTCAG, and 1492R, CGGTTACCTTGTTACGACTT. The results were compared with the 16S rRNA sequence available in public nucleotide databases at the EzBioCloud by using its World Wide Web site (http://www.ezbiocloud.net) and the basic local alignment search tool (BLAST) algorithm. Multiple sequence alignment and phylogenetic analysis were performed by the Neighbour-joining (NJ) method embedded in the software package MEGA (version 6).
The presence of the following enzymes was analyzed: amylase, lipase, and protease. The pure cultures of isolates were inoculated onto solid diagnostic media and were incubated at screening temperature for 24-48 h. Amylase activity was determined in Marine agar medium with 0.2% soluble starch (BD, Sparks, MD, USA). The plates were observed for clear zone near the inoculated colony after treatment with iodine solution. Marine agar medium supplemented with 2% skim milk (BD, Sparks, MD, USA) was prepared as a protease substrate. Observation of the transparent zone indicated positive protease activity. The strains were inoculated onto the plates with 1% Tween 80 (Sigma-Aldrich, Burlington, MA, USA). White precipitation around the boundary of the colony was indicative of lipase activity.
Results and Discussion
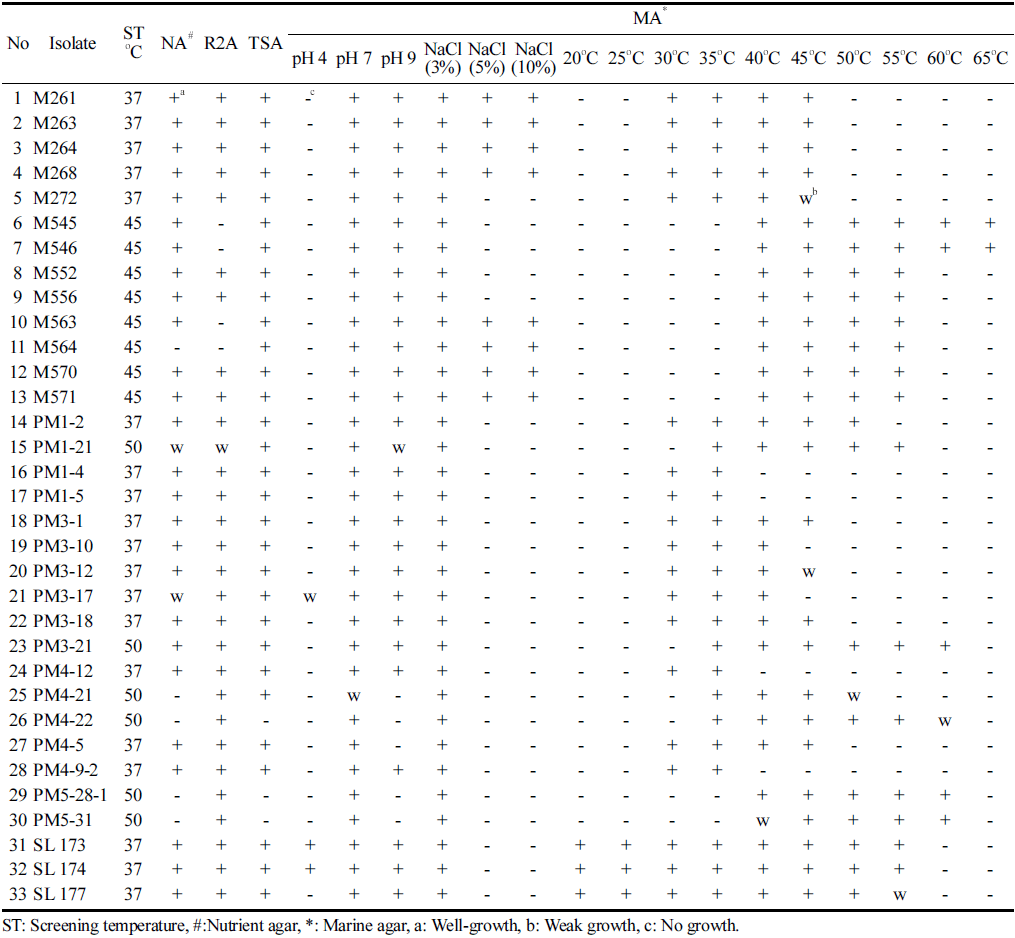
Within the scope of this study, a total of 33 strains were isolated from arable soil (Silla Garden, Busan, South Korea) fertilized with peat moss from West Siberia. Screening results showed that not only mesophiles were isolated but also strains of thermophilic bacteria were isolated from soil samples. The temperature, pH, and salinity tolerance profile of the strains are shown in Table 1. The temperature profile of the isolates indicated that 18 strains grow at temperatures higher than 45 ° C. Among them, 6 strains (PM3-21, PM5-28-1, PM5-31, PM4-22, M545, and M546) showed growth at 60 ° C with 2 strains (M545, M546) growing at 65 ° C. These results indicated that these strains are thermophilic bacteria. On the other hand, 15 strains were mesophiles, with 9 strains (M261, M263, M264, M268, M272, PM3-1, PM3-12, PM3-18, PM4-5) showing growth at 45 ° C, indicating that they are thermotolerant mesophilic bacteria. Among them, isolate M264 was identified as Bacillus subtilis meaning that the thermotolerant strain of the mesophilic bacterium Bacillus subtilis was isolated through this study. Thermophilic microorganisms that are capable of living at low as well as high temperatures is an attention-grabbing group (Rothschild and Mancinelli, 2001). 3 strains (SL 177, SL 173, and SL 174) showed broad temperature range growth from 20 ° C to 55 ° C. Furthermore, the strains SL 173, and SL 174 showed growth in both acidic and alkaline conditions. Generally, the majority of the strains (84.8%) could grow in alkaline condition. All strains could tolerate salinity up to 3% NaCl (w/v) concentration, and only 8 isolates could tolerate up to 5-10%. These results indicate that all isolates are halotolerant bacteria, besides the 8 isolates that are halophiles. Tolerance to a wide range of pH and moderate salt concentration are other important characteristics of these bacterial isolates besides temperature tolerance. In addition to this, all strains were able to grow in at least one complex medium (including weak growth) confirming the potential industrial application of the isolated strains.
|
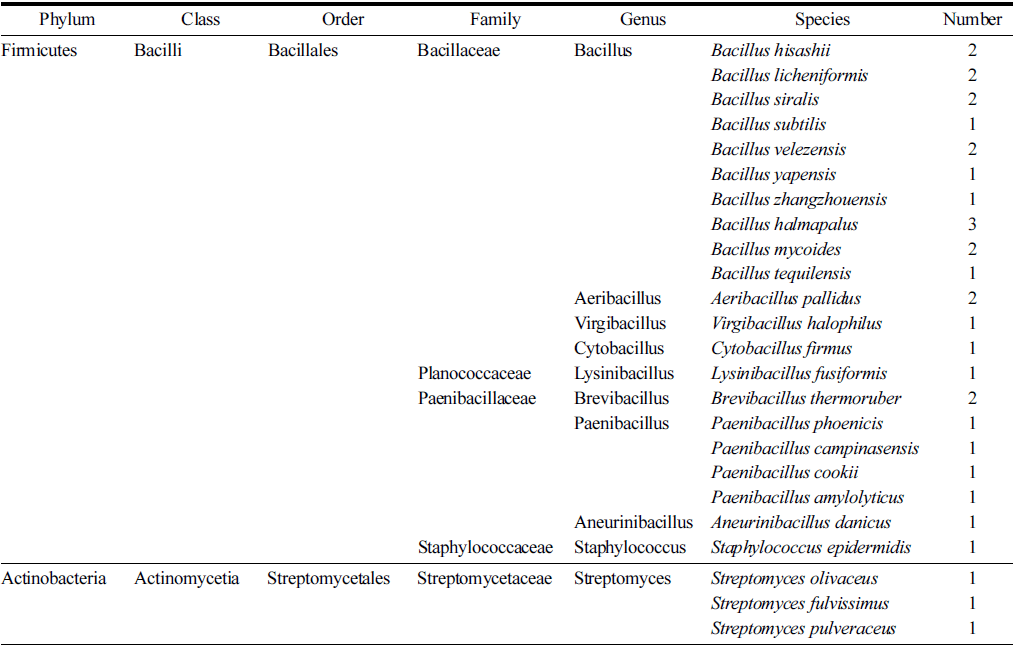
The 16S rRNA gene amplicon sequencing of all isolates was successfully achieved. As a result of pairwise comparisons of the 16S rRNA gene sequences on EzBioCloud server, 24 species were identified. All strains were divided into 2 phyla, 5 families, 10 genera, and 24 species (Table 2) and the identification of strains and their top-hit reference species is shown in Table 3. Identified strains belonged to phylum Firmicutes and Actinobacteria. 30 isolates belonged to phylum Firmicutes, class Bacilli and were divided into 4 families. Strains belonging to the family Bacillaceae were dominant with 17 strains of Bacillus sp., 2 strains of Aeribacillus sp., 1 strain of Virgibacillus sp., and 1 strain of Cytobacillus sp. The strains of Paenibacillaceae family were divided into 3 genera with 2 strains of Brevibacillus sp., 4 strains of Paenibacillus sp., and 1 strain of Aneurinibacillus sp. Strains of family Planococcaceae and Staphylococcaceae were less dominant with 1 strain of Lysinibacillus sp. and 1 strain of Staphylococcus sp., respectively. As for the phylum Actinobacteria, 3 strains of Streptomyces sp. were identified. The abundance of Bacillus and Paenibacillus species among the isolates can be attributed to their survival ability and endospore formation (Nicholson et al. 2000). On the other hand, Bacilli class is one of the dominant taxonomic groups in the bacterial complex of peat of Western Siberia (Golovchenko et al., 2004). Phylogenetic analysis was conducted to evaluate the relationships of the isolated strains. The results of the phylogenetic analysis (Fig. 1) of the 16S rRNA sequences showed that isolates represented a wide diversity, with the bacterial genera Bacillus, Aeribacillus, Virgibacillus, Cytobacillus, Lysinibacillus, Brevibacillus, Paenibacillus, Aneurinibacillus, Staphylococcus, and Streptomyces.
|
|

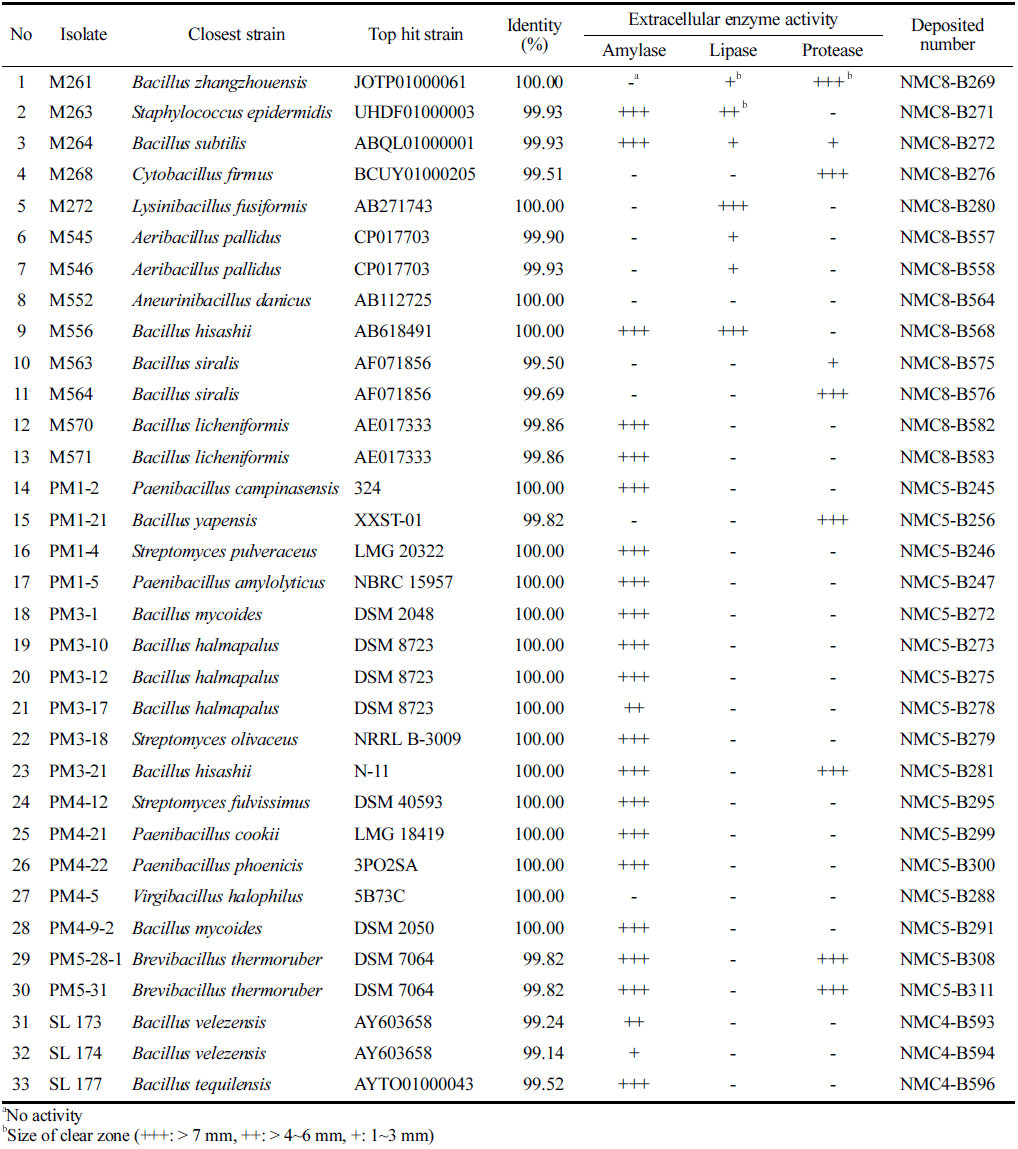
In order to confirm the industrial applicability of the isolated strains, industrially valuable enzymes such as amylase, lipase, and protease activity tests were performed. After the enzyme activity assay was completed, it was confirmed that all isolates have at least one hydrolase activity, except the isolate PM4-5. Amylase activity was detected in 24, lipase activity in 7, protease activity in 9 strains. Amylolytic activity was the most common among isolates with highest amylase activity in 21 strains. Highest protease activity was detected in 7 strains and highest lipase activity was detected in 2 strains M556 and M272. Combined hydrolytic activities were detected in 7 strains. To be detailed, both amylolytic and lipolytic activity was detected in 2 strains that were identified as Bacillus hisashii M556 and Staphylococcusepidermidis M263. Halophilic strain M261 was able to produce both lipase and protease enzymes. 3 strains (PM3-21, PM5-28-1, and PM5- 31) that grow at 60 ° C showed both proteolytic and amylolytic activity. Moreover, strain M264 that was able to grow at 45 ° C and could tolerate NaCl concentration (w/v) up to 10% showed all tested hydrolytic enzyme activity.
In conclusion, mesophilic and thermophilic strains isolated through this study can be utilized as basic biological material for hydrolytic enzyme-related applications in several industrial areas. Furthermore, this study is expected to have great significance in terms of diversity of domestic microbial biological resources and expanding scientific knowledge on the diversity and characteristics of fertilized soil bacteria. Also, all strains isolated through this study were deposited in the Microbial Value Enhancement Project, Korea Research Institute of Bioscience and Biotechnology.